As several historians of the contemporary life sciences have pointed out, much of biological thinking is metaphorical. Not least in genomics. The race for the human genome in the 1990s and much of subsequent postgenomics has been guided by a series of metaphors that draw on linguistics, management science, and information and communication science.
Accordingly, computer science analogues have flourished. The genome is said to contain the ‘code’ or ‘blueprint’ for development, the ribosomes ‘read’ the instructions, and ‘information’ is carried on to proteins through ‘transcription’ and ‘translation’ (see, for example, Susanne Knudsen’s analysis of the ‘code’ metaphor in ‘Communicating novel and conventional scientific metaphors: a study of the development of the metaphor of genetic code’ (Publ. Commun. Sci., vol 14, pp. 373-92, 2005).
 But linguistic and computer analogies are not very helpful when it comes to the proteome with its estimated half million or so specific protein-protein interactions (the ‘interactome‘). What kinds of metaphors are available if one wants to capture this rich and complex protein universe?
But linguistic and computer analogies are not very helpful when it comes to the proteome with its estimated half million or so specific protein-protein interactions (the ‘interactome‘). What kinds of metaphors are available if one wants to capture this rich and complex protein universe?
This is not just an academic question, but also a practical one: metaphors are indispensible tools for science communicators. So reflective answers to this question would be very useful for science and medicine museums that wish to present protein science and proteomics in their displays — as we are planning to do in a small extramural exhibition next year.
Browsing the literature gives a few tentative hints. Proteins are usually spoken about in quite mundane metaphorical terms, a far cry from the informational control center terminology that guided the rise of genomics. Thus the only major historical overview in book form so far, written by now retired protein researchers Charles Tanford and Jacqueline Reynolds (Oxford UP, 2001), is aptly titled Nature’s Robots.
Others — for example this University of Washington site — liken proteins to the ‘work-horses’ of the life processes. The two images reproduced in this post also suggest metaphors that gesture towards a mechanical, workman-filled world. The small image above illustrates a proposed ‘cogwheel model’ for signal transduction across membranes, involving a transmembrane protein receptor in the form of a four-helical coil. The model was on the cover of Cell (8 September, 2006), explained as “a gear box with four cogwheels” (credit: Martin Voetsch, Max Planck Institute for Developmental Biology).
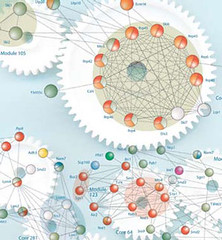 The second image is also a ‘cogwheel model’, in this case of a protein complex in yeast cytoplasm that contributes to mRNA decay (credit: European Molecular Biology Laboratory; from The Scientist 22, issue 9, p. 55, 2008)
The second image is also a ‘cogwheel model’, in this case of a protein complex in yeast cytoplasm that contributes to mRNA decay (credit: European Molecular Biology Laboratory; from The Scientist 22, issue 9, p. 55, 2008)
There are other interesting metaphors around. University of Texas oncologist Gus Pappas has likened the proteome to ‘a play’s cast’, a list of the ‘Dramatis Personae’ of the cell (see ‘A new literary metaphor for the genome or proteome’, Biochemistry and Molecular Biology Education, vol. 33 (1), p. 15, 2006).
Maybe one could try further along these lines. Not in terms of literary or theatrical metaphors though, but in terms of politics. For example, imagine the cell as a city like Fritz Lang’s Metropolis, where a small elite caste of nucleic acids stand against a diverse multitude of life-producing worker-proteins — a proteomic metaphor that reflects some of the basic aspects of capitalism.